Sequences Analysed
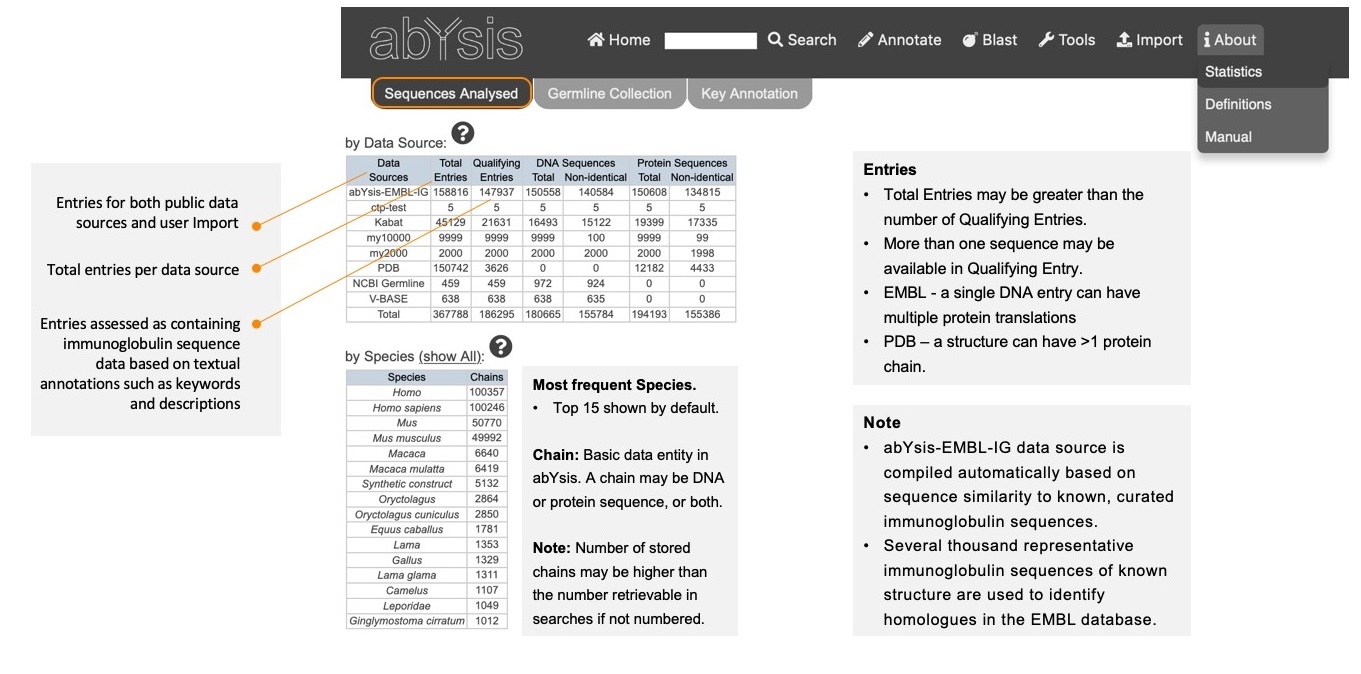
by Data Source
Data Sources Entries from public resources or Import function.
Total Entries Total number of entries for each Data Source - including non-antibodies.
Qualifying Entries Those assessed as containing immunoglobulin sequence data based on textual annotations such as keywords and descriptions.
More than one sequence may be available in a Qualifying
Entry so total number of sequences may be greater than the number
of qualifying entries.
Note: abYsis-EMBL-IG data source is compiled automatically based on sequence similarity to known, curated immunoglobulin sequences.
- Several thousand representative immunoglobulin query sequences of known structure have been used to identify immunoglobulins in the EMBL database whilst also minimising mis-selection of T-cell receptors.
by Species
Most frequent number of entries per species in abYsis. Top 15 Shown by default.
Note: Number of stored chains may be higher than the number retrievable in searches.
Chain: Basic data entity in abYsis. A chain may be DNA or protein sequence, or both.